Abstract
Background: Monosomy 7 (-7) is a typical abnormality in MDS with a frequency of ~ 10%. -7 either occurs as the sole abnormality (~4%), accompanied by one additional abnormality (~1%) or in a complex karyotype (~5%). A detailed molecular analysis of MDS with -7 as the sole abnormality (-7 sole) has not been performed to date.
Aims: Evaluation of molecular mutations in MDS with -7 sole and analysis of the clonal hierarchy.
Patient cohorts and methods: 100 patients (pts) with MDS at diagnosis harboring -7 sole in chromosome banding analysis (59 male, 41 female) with available cytomorphology and material for mutation screening were selected. Median age was 70 years (range: 21-89 years). 86 pts had de novo and 14 pts therapy-related MDS. Blast count was <5% in 29 MDS and ≥5% in 71 MDS pts. 97 pts were also analyzed by interphase FISH using a probe for the centromeric region of chromosome 7. Targeted sequencing of 29 genes commonly mutated in myeloid malignancies was performed in all 100 pts (Illumina, San Diego, CA). Variants of unknown significance were excluded from statistical analysis.
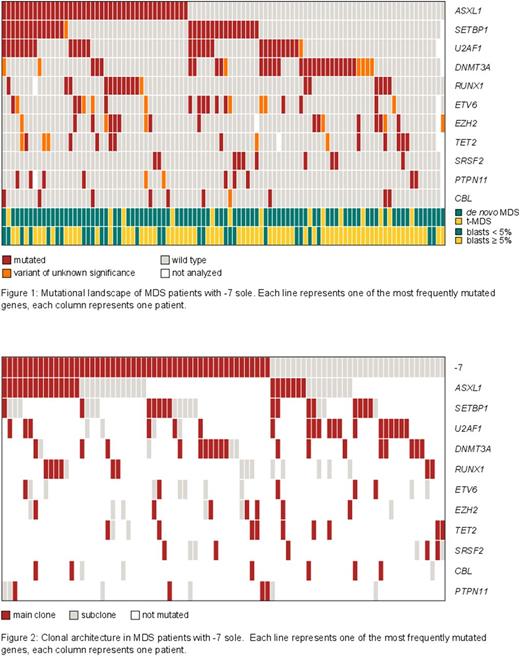
Results: The most frequently mutated genes were: ASXL1 (42%), SETBP1 (31%), U2AF1 (29%), DNTM3A (27%), RUNX1 (17%), ETV6 (14%), EZH2 (14%), TET2 (12%), SRSF2 (10%), CBL (9%), PTPN11 (9%), NF1 (6%), CSF3R (4%), JAK2 (4%), TP53 (3%), KRAS (3%), NRAS (3%), and MPL (3%). Genes mutated in 2% or less of pts were: GATA2, IDH1, IDH2, KIT, CALR, ETNK1, NPM1, SF3B1, and WT1 . No mutations were observed in CSNK1A1 and GATA1 (figure 1).
RUNX1 mutations were significantly more frequent in t-MDS as compared to de novo MDS (43% vs 13%, p=0.02), while ASXL1 mutations were more frequent in MDS with blast count <5% (60% vs 41%, p=0.04).
A median of 2 mutations per case were observed (range 0-6), with 94 pts harboring at least 1 mutation. No association between age and the number of mutations was observed. However, mutations in ETV6, SETBP1, U2AF1 and PTPN11 were associated with a higher number of mutations, while this was not the case for other gene mutations. In pts with 0-2 mutations ETV6, SETBP1, U2AF1 and PTPN11 were mutated in 4%, 15%, 14%, and 2%, respectively, while the respective mutation frequencies were 27%, 49%, 47%, and 19% in pts with >2 mutations (for all comparisons p<0.01).
The DNMT3A mutation frequency was low in ASXL1 mutated pts as compared to ASXL1 wild type (wt) pts (8% vs 42%; p<0.0001), while RUNX1 mutations were more frequent in ASXL1 mutated pts (28% vs 10%, p=0.03). U2AF1 and ETV6 mutations frequently co-occurred as 67% of ETV6 mutated pts also harboured a U2AF1 mutation (p=0.002) while only 21% of ETV6 wt pts showed U2AF1 mutations. In addition, the following mutation combinations were more frequent than expected U2AF1 / ETV6 (p=0.002), RUNX1 / EZH2 (p=0.04), EZH2 / TET2 (p=0.04), SETBP1 / U2AF1 (p=0.01), SETBP1 / PTPN11 (p=0.02), and U2AF1 / TP53 (p=0.02), while the following mutation combinations co-occurred less frequently than expected: EZH2 / SETPB1 (p=0.05), RUNX1 / SETBP1 (p=0.02), and SRSF2 / U2AF1 (p=0.02).
For clonal architecture analysis the clone size of cells harboring -7 was related to the mutation loads of the 11 most frequently mutated genes. The proportion of cells harboring -7 determined by FISH analyses varied between 6% and 95% (median: 57%). The -7 clone size was significantly correlated to the mutation loads of DNMT3A (r=0.67, p<0.001), RUNX1 (r=0.65, p=0.003), PTPN11 (r=0.71, p=0.02), and NF1 (r=0.81, p=0.01) indicating that -7 and the respective mutation were present together in the same clone. The mutation loads of the following mutations were significantly correlated: ASXL1 / PTPN11 (r=0.95, p=0.01), ASXL1 / U2AF1 (r=0.58, p=0.04), ETV6 / SETBP1 (r=0.91, p=0.005), and ETV6 / U2AF1 (r=0.87, p=0.03). Estimated based on clone size and mutation load -7 was present in the main clone (MC) in 61% of cases and in the subclone (SC) in 39%. Most mutations occurred in the MC as well as in the SC at comparable frequencies (ASXL1, EZH2, RUNX1, SETBP1, SRSF2). Mutations in ETV6 and PTPN11 were present slightly more frequently in a SC only (64% and 60%), while mutations in CBL, DNMT3A, TET2 and U2AF1 were found rarely in a SC only (33%, 18%, 18% and 17%) (figure 2).
Conclusions: A broad spectrum of molecular mutations is present in MDS with -7 sole with the highest mutation frequencies in ASXL1, SETBP1, U2AF1, DNMT3A and RUNX1 . No clear hierarchy of -7 and mutations was identified, however, in most cases if mutated DNMT3A, TET2 and U2AF1 are present in the main clone.
Haferlach: MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Walter: MLL Munich Leukemia Laboratory: Employment. Stengel: MLL Munich Leukemia Laboratory: Employment. Nadarajah: MLL Munich Leukemia Laboratory: Employment. Kern: MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Meggendorfer: MLL Munich Leukemia Laboratory: Employment. Haferlach: MLL Munich Leukemia Laboratory: Employment, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal